|
品牌 |
酶研生物 | |||||||||||
货号 |
MY1637 | |||||||||||
规格 |
2ug | |||||||||||
价格 |
询价 | |||||||||||
货期 |
现货 | |||||||||||
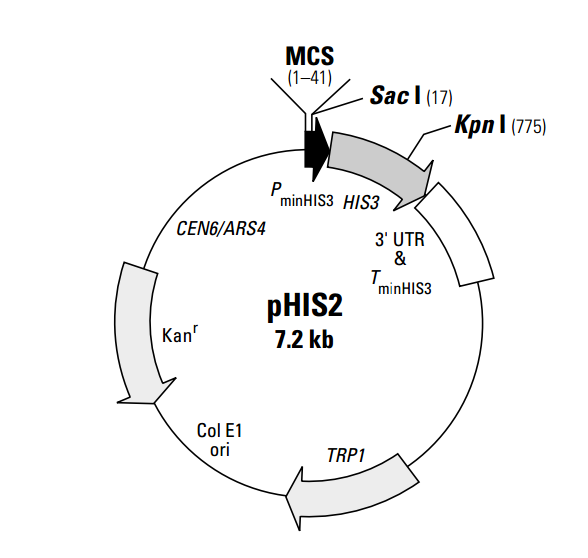
pHIS2 is a reporter vector that can be used in yeast one-hybrid assays to identify and characterize DNA-binding proteins. The vector was specifically designed for use with the BD Matchmaker™ One- Hybrid Library Construction & Screening Kit (#K1617-1). It contains a HIS3 nutritional reporter gene, located downstream of a multiple cloning site (MCS) and the minimal promoter of the HIS3 locus (PminHIS3). Cis-acting DNA sequences, or DNA target elements, can be inserted into the MCS and used as baits to screen GAL4 AD/cDNA fusion libraries for proteins that interact with the target sequence. A protein-DNA (or one-hybrid) interaction can be detected by performing the assay in a yeast strain such as Y187 that is auxotrophic for histidine. Positive one-hybrid interactions drive expression of the HIS3 reporter gene, which enables the host cell to grow on histidine-deficient media. In the absence of activation, the constitutive HIS3 expression from PminHIS3 is very low. During library screening, the leaky expression of HIS3 is controlled by adding 3-amino-1,2,4-triazole (3-AT) to the medium. The concentration of 3-AT needed to fully suppress leaky HIS3 expression must be determined empirically for each DNA target element. pHIS2 can be maintained in both yeast and bacteria. It contains an autonomous replication sequence (ARS4) and TRP1 nutritional marker for replication and selection in yeast (1, 2); it contains a Col E1 origin and a kanamycin resistance gene (Kanr) for propagation and selection in E. coli. The centromeric sequence CEN6 ensures proper segregation of the plasmid during cell division in yeast (1, 2). 载体应用To use pHIS2 in a one-hybrid assay, clone one or more copies of a cis-acting DNA target sequence into the MCS. Then introduce the plasmid into competent yeast cells using the transformation protocol in the BD Matchmaker Library Construction & Screening Kits User Manual (PT3529-1). In contrast to the original BD Matchmaker One- Hybrid System, this reporter vector does not need to be integrated into the yeast genome. Instead, it is maintained as an episome throughout the assay. Inserting your target element may alter the level of background HIS3 expression. Therefore, constructs should be tested for background (leaky) HIS3 expression before you start a one-hybrid analysis. Background growth due to leaky HIS3 expression is controlled by adding 3-AT to the selection medium, as described in the User Manual (PT3529-1).
|
|
|||||||||||